Projects
Our lab uses systems biology to understand how dysfunctional signaling states drive cancer heterogeneity and therapeutic response. By integrating multi-omics data with computational and experimental models, we identify mechanisms and vulnerabilities that enable precision cancer treatment. Current projects include:
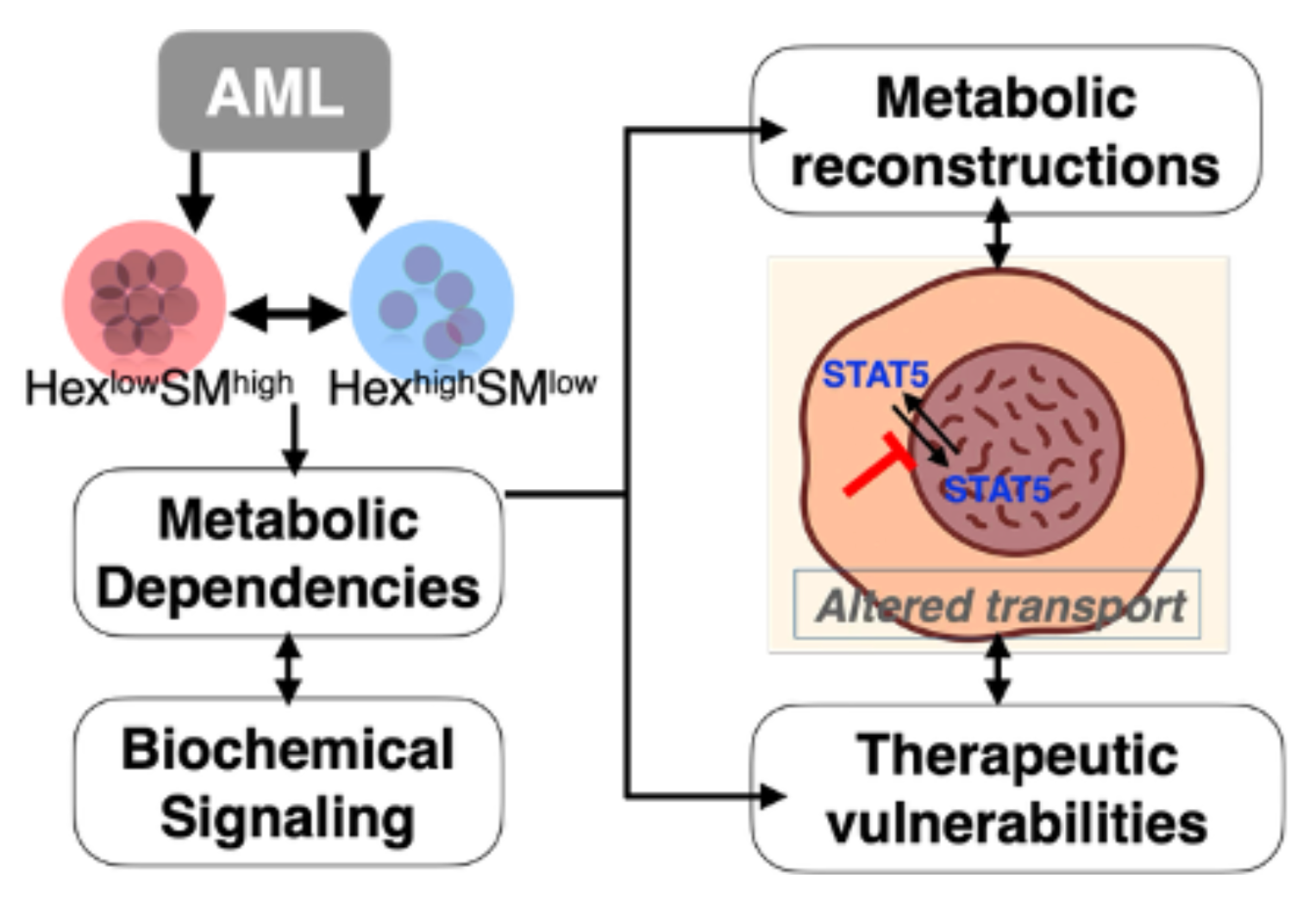
- Defining mechanistic basis for metabolic heterogeneity in tumors
- Identifying therapeutic vulnerabilities and risk-classification
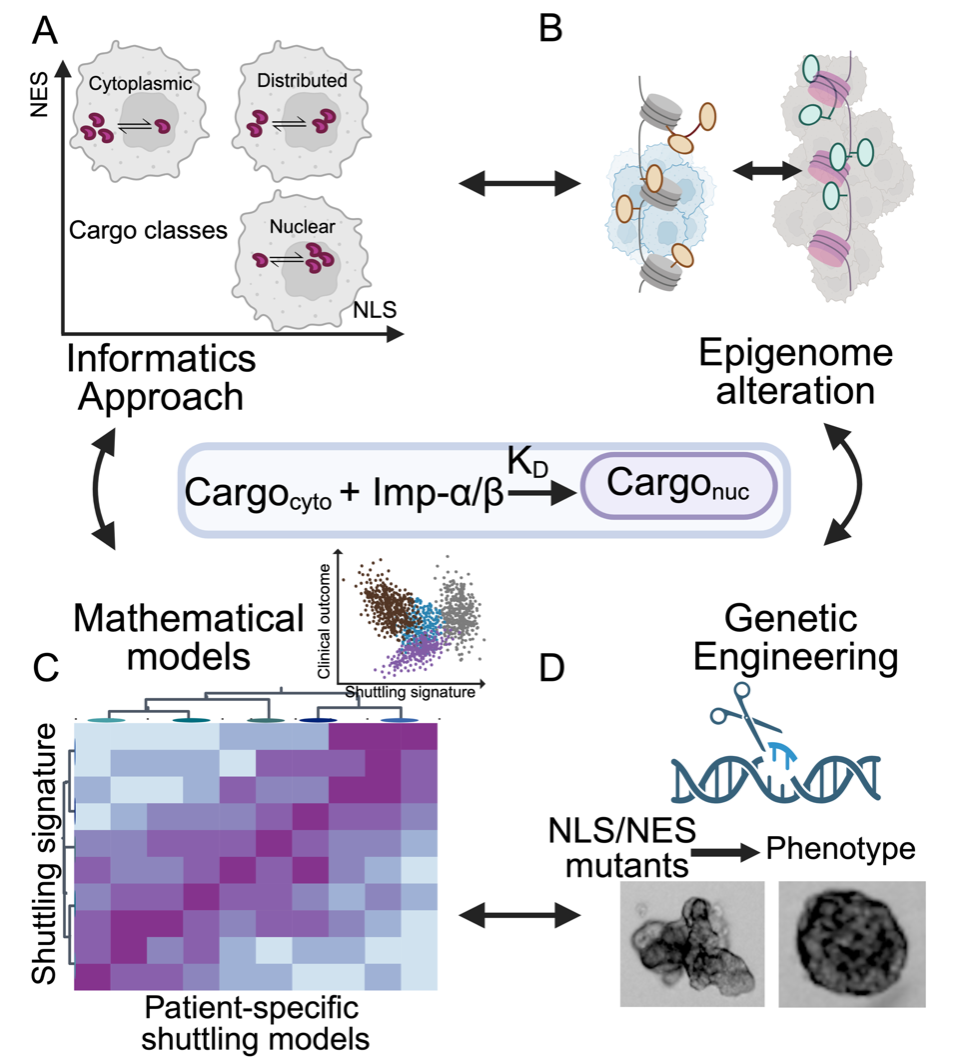
- Determining links between altered epigenome and tumorigenesis
- Systems-level investigation of biomarkers for health disparities
This page highlights software tools, datasets, and research-driven resources developed to support data analysis, reproducibility, and integrative modeling. The projects span methodological development, computational workflows, and applied analyses, with an emphasis on transparency, usability, and impact across biomedical and quantitative research. We welcome feedback, comments on the usability of these tools.
Featured
More

This tool will allow you to visualize your CRISPR screen data once you have the count table and gene summary table obtained by Mageck.

SVM classifier to predict HexSM lipid subtype using normalized gene expression data

List of available datasets for AML within NCI funded P01 within UVA/PSU/Wake/MSKCC

Developed by Janes lab to examine relationship between mRNA and protein.